Pseudogene
Pseudogenes are genes that have lost their function. They have lost their gene expression in the cell or their ability to code protein.[1] The term was coined in 1977.[2]
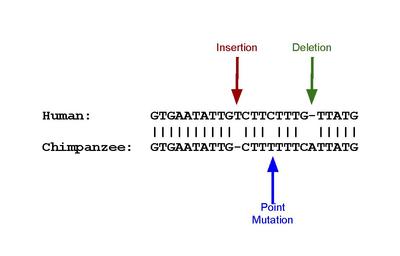
Pseudogenes can result from mutations in a gene whose product is not needed for the survival of the organism. Although not protein-coding, the DNA of pseudogenes may be functional.[3] It may be similar to other kinds of non-coding DNA which have a regulatory role.
Most have some gene-like features. They lack protein-coding ability resulting from a variety of disabling mutations, or their inability to encode RNA (such as with rRNA pseudogenes).[4]
Pseudogenes are generally thought of as the last stop for genomic material that is to be removed from the genome,[5] so they are often labeled as junk DNA. Pseudogenes contain fascinating biological and evolutionary histories in their sequences. This is due to a pseudogene's shared ancestry with a functional gene. In the same way that Darwin thought of two species as having a shared common ancestry followed by millions of years of evolutionary divergence (see speciation), a pseudogene and its associated functional gene also share a common ancestor and have diverged as separate genetic entities over millions of years.
References
change- ↑ Vanin EF (1985). "Processed pseudogenes: characteristics and evolution". Annu. Rev. Genet. 19: 253–72. doi:10.1146/annurev.ge.19.120185.001345. PMID 3909943.
- ↑ Jacq C; Miller J.R. Brownlee G.G. (September 1977). "A pseudogene structure in 5S DNA of Xenopus laevis". Cell. 12 (1): 109–20. doi:10.1016/0092-8674(77)90189-1. PMID 561661. S2CID 25965491.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Poliseno L (2010). "A coding-independent function of gene and pseudogene mRNAs regulates tumour biology". Nature. 465 (7301): 1033–1038. Bibcode:2010Natur.465.1033P. doi:10.1038/nature09144. PMC 3206313. PMID 20577206.
- ↑ Herron, Jon C; Freeman, Scott (2007). Evolutionary analysis. Upper Saddle River, NJ: 4th ed, Pearson Prentice Hall. ISBN 978-0-13-227584-2.
{{cite book}}: CS1 maint: multiple names: authors list (link) [1] - ↑ Zheng D. et al 2007. Pseudogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Res. 17 (6): 839–51. [2]