ZNF839
| ZNF839 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ZNF839, C14orf131, zinc finger protein 839 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | MGI: 1920055; HomoloGene: 49541; GeneCards: ZNF839; OMA:ZNF839 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
ZNF839 or zinc finger protein 839 is a protein which in humans is encoded by the ZNF839 gene. It is located on the long arm of chromosome 14.[5] Zinc finger protein 839 is speculated to play a role in humoral immune response to cancer as a renal carcinoma antigen (NY-REN-50). This is because NY-REN-50 was found to be over expressed in cancer patients, especially those with renal carcinoma.[6][7] Zinc finger protein 839 also plays a role in transcription regulation by metal-ion binding since it binds to DNA via C2H2-type zinc finger repeats.[8][9]
Gene
[edit]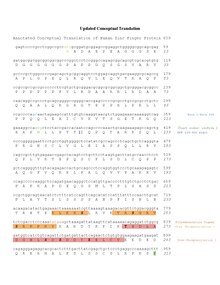
The human ZNF839 gene is 25,326 nucleotides long and it encodes transcript variant 1 which is the longest isoform.[10] The gene locus is 14q32.31 and it is found on the plus strand.[10] ZNF839 consists of 8 exons, it encodes for one zinc finger repeat, and 3 disordered regions.[9] The transcript variant 2 (isoform 2) is missing exon 1 so it has a downstream start codon.
mRNA/transcript
[edit]The mRNA isoform 1 encoded by the human ZNF839 gene is 2992 nucleotides long. There are 9 isoforms of the ZNF839 mRNA. [11]
| Isoform Number | Accession Number | mRNA Length (nt) | Protein Length (AA) | Molecular Weight (kDa) |
|---|---|---|---|---|
| 1 | NM_018335 | 2992 | 927 | 87.5 |
| 2 | NM_001267827 | 2845 | 811 | 87.4 |
| 3 | NM_001385065 | 2842 | 877 | 93.5 |
| 4 | NM_001385069 | 3017 | 806 | 86.9 |
| 5 | NM_001385070 | 2695 | 761 | 81.5 |
| 6 | NM_001385071 | 2620 | 736 | 79.3 |
| 7 | NM_001385072 | 2212 | 667 | 72.4 |
| 8 | NM_001385073 | 2062 | 617 | 66.6 |
| 9 | NM_001385076 | 2065 | 551 | 60.5 |
Expression
[edit]It has been found that there is high expression of ZNF839 in the testis, specifically the seminiferous tubules, after RNA-sequencing analysis of various human tissues.[11][12][13] The high expression of the human ZNF839 gene in the testis may be due to its C2H2-zinc finger role in transcriptional regulation during spermatogenesis.[14] Thus, zinc finger protein 839 role's as a transcription factor may be essential in regulating spermatogenesis. There is also high expression of zinc finger protein 839 in the various regions of the gyri in the brain.[15] The high expression of ZNF839 in the gyri of the cerebrum may be due to the regulatory involvement of zinc finger proteins in neurodevelopment.[16]

Protein
[edit]The human zinc finger protein 839, isoform 1, has 927 amino acids and holds a molecular weight of 87.5 kDa. The predicted isoelectric point of zinc finger protein 839 is 6.18.[17] This protein contains one zinc finger and one domain of unknown function (DUF) between amino acids 123-290. In isoforms 2-9, there is variability in the location of the DUF. The C2H2 zinc fingers in zinc finger protein 839 consist of cysteine and histidine residues that are conserved.[18]
Subcellular localization
[edit]Zinc finger protein 839 is predicted to be localized to the nucleus due to its role as a transcription factor.[19][20] The pat4 nuclear localization signal was conserved among all strict orthologs of the human zinc finger protein 839.

Tertiary structure
[edit]The human zinc finger protein 839 tertiary structure is predicted to have 5 alpha helices, one beta-sheet, and the rest of the protein consists of coils. Two of the alpha helices were found in two phosphorylation sites. The prevalence of negative and positive charges is mostly found in the coils.
Function
[edit]Interacting proteins
[edit]Zinc finger protein 839 was found to have the following interacting proteins: TP53 (Tumor Protein p53), YWHAE (14-3-3 Protein Epsilon), YWHAZ (Tyrosine-3-monooxygenase), APP (amyloid beta precursor protein).[21] It was found that YWHAE is involved in signal transduction pathways due to its binding of phosphoserine-containing proteins.[22] Zinc finger protein 839 has many phosphorylation sites on serine residues since phosphorylation is one of the significant post-translational modifications that was found in ZNF839. Thus, YWHAE is able to interact with zinc finger protein 839 since it has multiple phosphoserines after post-translational modification. In addition, it was found that when a C2H2-zinc finger and p53 DNA binding domain fusion transcription factor was created, it increased the transcription of gene downstream of p53 such as p21, whose function is to arrest cell cycle in response to DNA damage.[23] Thus, TP53 and zinc finger protein 839 may interact to transcribe genes necessary for DNA damage response or tumor suppression. Overall, it was observed that there is a pattern of zinc finger protein 839 interactions with tumor suppressor proteins and signal transduction regulators. Thus, zinc finger protein may significantly play a role in tumor suppression and regulation of cellular pathways such as cell division.
Clinical significance
[edit]A relationship has been found between colorectal cancer patient survival and human ZNF839 expression.[24] The appearance of rs11704 single nucleotide polymorphism (SNP) in the miRNA binding site of the 3’ UTR within ZNF839 mRNA resulted in loss of miRNA binding site and leads to up-regulation of the human ZNF839 gene.[6] The up-regulation of the ZNF839 gene results in decreased survival of colorectal cancer patients.[24]
Post-translational modifications
[edit]Zinc finger protein 839 has five phosphorylation sites and one sumoylation site.[25][26] The kinases involved in the phosphorylation of zinc finger protein 839 include: Protein Kinase C, Casein Kinase II, and Casein Kinase I. These kinases play a role in activating transcription of genes involved in DNA repair, cell growth, and cell proliferation.

Evolution
[edit]Orthologs
[edit]Zinc finger protein 839 is found only in vertebrates, not invertebrates. Zinc finger protein 839 is found in the following vertebrates: mammals, birds, reptiles, amphibians, and fish. Zinc Finger Protein 839 was approximately found to have first appeared in fish 462 million years ago. The sequence length for Japanese quail, white-collared manakin, lanner falcon, atlantic canary, central bearded dragon, and aeolian wall lizard have been trimmed for an accurate sequence identity.
| ZNF839 | Genus | Common Name | Taxonomic Group | Date of Divergence (MYA) | Accession Number | Sequence Length (aa) | Sequence Similarity (%) | Sequence Identity(%) |
|---|---|---|---|---|---|---|---|---|
| Mammal | Homo sapiens | Human | Primates | 0 | NP_060805 | 927 | 100 | 100 |
| Mus musculus | Mouse | Rodentia | 87 | NP_082641 | 921 | 61.5 | 49.8 | |
| Hipposideros armiger | Great roundleaf bat | Chiroptera | 94 | XP_019494952 | 916 | 60.9 | 53.2 | |
| Elephas maximus indicus | Indian Elephant | Proboscidea | 99 | XP_049755600 | 937 | 61.1 | 52.8 | |
| Choloepus didactylus | Linneaus’s two-toed sloth | Pilosa | 99 | XP_037687739 | 940 | 60.4 | 52.5 | |
| Aves | Falco biarmicus | Lanner Falcon | Falconiformes | 319 | XP_056201354 | 1010 | 47.6 | 34.5 |
| Serinus canaria | Atlantic canary | Passeriformes | 319 | XP_030095548 | 974 | 46.7 | 35 | |
| Manacus Candei | White-collared manakin | Passeriformes | 319 | XP_051625574 | 980 | 46.7 | 34.8 | |
| Coturnix japonica | Japanese quail | Galliformes | 319 | XP_015720921 | 931 | 44.3 | 33.2 | |
| Reptilia | Podarcis raffonei | Aeolian wall lizard | Squamata | 319 | XP_053226315 | 776 | 37.8 | 26.8 |
| Pogona vitticeps | Central bearded dragon | Squamata | 319 | XP_020649269 | 669 | 37.6 | 27.3 | |
| Zootoca vivpara | Viviparous lizard | Squamata | 319 | XP_034961572 | 777 | 35.9 | 26.8 | |
| Sceloporus undulatus | Eastern Fence Lizard | Squamata | 319 | XP_042302042 | 696 | 35.8 | 25.1 | |
| Amphibians | Microcaecilia unicolor | Tiny cayenne caecillian | Gymnophiona | 352 | XP_030070400 | 1133 | 42.1 | 29.5 |
| Geotrypetes seraphini | Gaboon caecillian | Gymnophiona | 352 | XP_033808581 | 1126 | 41.4 | 29.2 | |
| Bombina bombina | European fire-bellied toad | Anura | 352 | XP_053553408 | 937 | 38.8 | 27.6 | |
| Rana temporaria | Common Frog | Anura | 352 | XP_040188931 | 965 | 35.4 | 22.8 | |
| Fish | Callorhinchus milii | Australian ghostshark | Chimaeriformes | 462 | XP_042194042 | 1247 | 35.3 | 24.6 |
| Leucoraja erinacea | Little skate | Rajiformes | 462 | XP_055497145 | 1256 | 34.9 | 23.3 | |
| Chiloscyllium plagiosum | Whitespotted bamboo shark | Orectolobiformes | 462 | XP_043553384 | 1269 | 33.9 | 22.9 | |
| Rhincodon typus | Whale shark | Orectolobiformes | 462 | XP_020377475 | 1265 | 33.1 | 23.3 |
Paralogs
[edit]Zinc finger protein 839 was found to have no paralogs.
Evolution Rate
[edit]In addition. the ZNF839 gene is evolving at a fast rate, due to its evolution rate being close to fibrinogen alpha chain's rate than cytochrome C.

References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000022976 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000021271 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "ZNF839 zinc finger protein 839 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2023-12-06.
- ^ a b Yang YP, Ting WC, Chen LM, Lu TL, Bao BY (2017-01-01). "Polymorphisms in MicroRNA Binding Sites Predict Colorectal Cancer Survival". International Journal of Medical Sciences. 14 (1): 53–57. doi:10.7150/ijms.17027. PMC 5278659. PMID 28138309.
- ^ Scanlan MJ, Gordan JD, Williamson B, Stockert E, Bander NH, Jongeneel V, et al. (November 1999). "Antigens recognized by autologous antibody in patients with renal-cell carcinoma". International Journal of Cancer. 83 (4): 456–464. doi:10.1002/(SICI)1097-0215(19991112)83:4<456::AID-IJC4>3.0.CO;2-5. PMID 10508479. S2CID 21839750.
- ^ "Alliance of Genome Resources". www.alliancegenome.org. Retrieved 2023-12-06.
- ^ a b "UniProt". www.uniprot.org. Retrieved 2023-12-06.
- ^ a b "Homo sapiens zinc finger protein 839 (ZNF839), transcript variant 1, mRNA". ncbi.nlm.nih.gov. 2022-12-29. Retrieved 2023-12-16.
- ^ a b "ZNF839 zinc finger protein 839 [Homo sapiens (human)] - Gene - NCBI". www.ncbi.nlm.nih.gov. Retrieved 2023-12-07.
- ^ Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. (February 2014). "Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics". Molecular & Cellular Proteomics. 13 (2): 397–406. doi:10.1074/mcp.m113.035600. PMC 3916642. PMID 24309898.
- ^ "GDS596 / 221709_s_at". www.ncbi.nlm.nih.gov. Retrieved 2023-12-07.
- ^ Ishizuka M, Ohtsuka E, Inoue A, Odaka M, Ohshima H, Tamura N, et al. (September 2016). "Abnormal spermatogenesis and male infertility in testicular zinc finger protein Zfp318-knockout mice". Development, Growth & Differentiation. 58 (7): 600–608. doi:10.1111/dgd.12301. PMID 27385512.
- ^ "Microarray Gene Detail :: Allen Brain Atlas: Human Brain". human.brain-map.org. Retrieved 2023-12-15.
- ^ Bu S, Lv Y, Liu Y, Qiao S, Wang H (2021). "Zinc Finger Proteins in Neuro-Related Diseases Progression". Frontiers in Neuroscience. 15: 760567. doi:10.3389/fnins.2021.760567. PMC 8637543. PMID 34867169.
- ^ "Expasy - Compute pI/Mw tool". web.expasy.org. Retrieved 2023-12-10.
- ^ Isalan M (2013-01-01), "Zinc Fingers", in Lennarz WJ, Lane MD (eds.), Encyclopedia of Biological Chemistry (Second Edition), Waltham: Academic Press, pp. 575–579, ISBN 978-0-12-378631-9, retrieved 2023-12-10
- ^ "PSORT II Prediction". psort.hgc.jp. Retrieved 2023-12-10.
- ^ "ZNF839 protein expression summary - The Human Protein Atlas". www.proteinatlas.org. Retrieved 2023-12-10.
- ^ "PSICQUIC View". www.ebi.ac.uk. Retrieved 2023-12-15.
- ^ "YWHAE", Wikipedia, 2023-08-18, retrieved 2023-12-13
- ^ Falke D, Fisher MH, Juliano RL (November 2004). "Selective transcription of p53 target genes by zinc finger-p53 DNA binding domain chimeras". Biochimica et Biophysica Acta. 1681 (1): 15–27. doi:10.1016/j.bbaexp.2004.09.011. PMID 15566940.
- ^ a b To KK, Tong CW, Wu M, Cho WC (July 2018). "MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside". World Journal of Gastroenterology. 24 (27): 2949–2973. doi:10.3748/wjg.v24.i27.2949. PMC 6054943. PMID 30038463.
- ^ "Frontpage". www.healthtech.dtu.dk. Retrieved 2023-12-15.
- ^ "GPS-SUMO: Prediction of SUMOylation Sites & SUMO-interaction Motifs". sumo.biocuckoo.cn. Retrieved 2023-12-15.